Biodiversity and the policy tools to manage it!
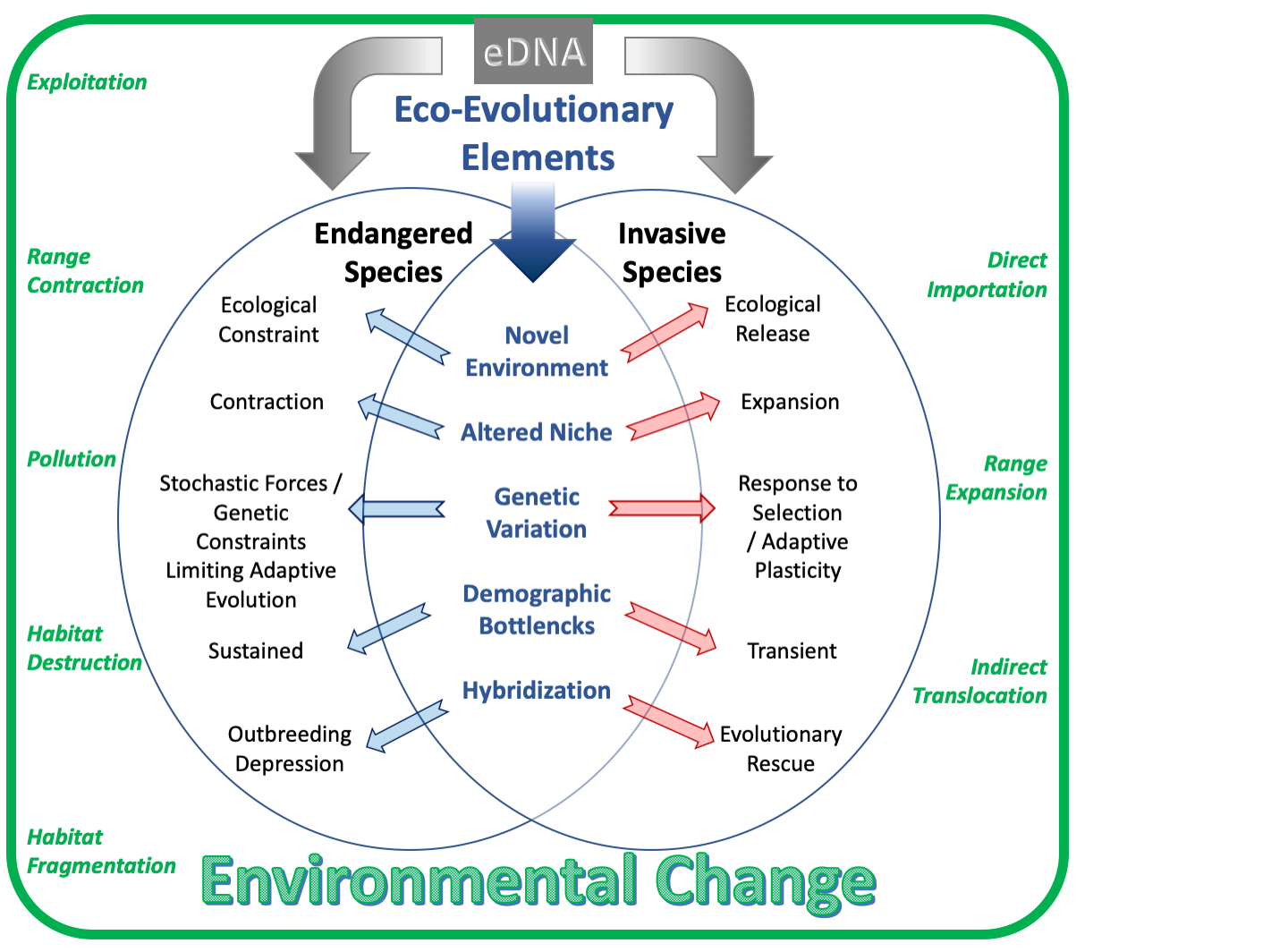
The Anthropocene has seen global biodiversity increasingly under threat from human activity, ultimately increasing both rates of extinction and invasion. In fact, the various eco-evolutionary challenges imposed along the full spectrum of species are similar in action yet different in their effect, offering complimentary research approaches. Thus, cradled within environmental change, I examine how eco-evolutionary elements affect different species, and use environmental DNA (eDNA) to monitor and manage them.
In short, I employ molecular tools to understand biodiversity:
(i) processes that elucidate diversification in changing environments, and (ii) practical applications for quantifying biodiversity and preventing its loss.
To accomplish these aims, I use field, lab and quantitative approaches, and various study systems (frogs, lizards, birds, mammals, and invertebrates) to lead a hypothesis-driven research program. Below are some of the major themes my research program investigates (click on the links above to read more).
(i) The origins and maintenance of diversity under environmental change
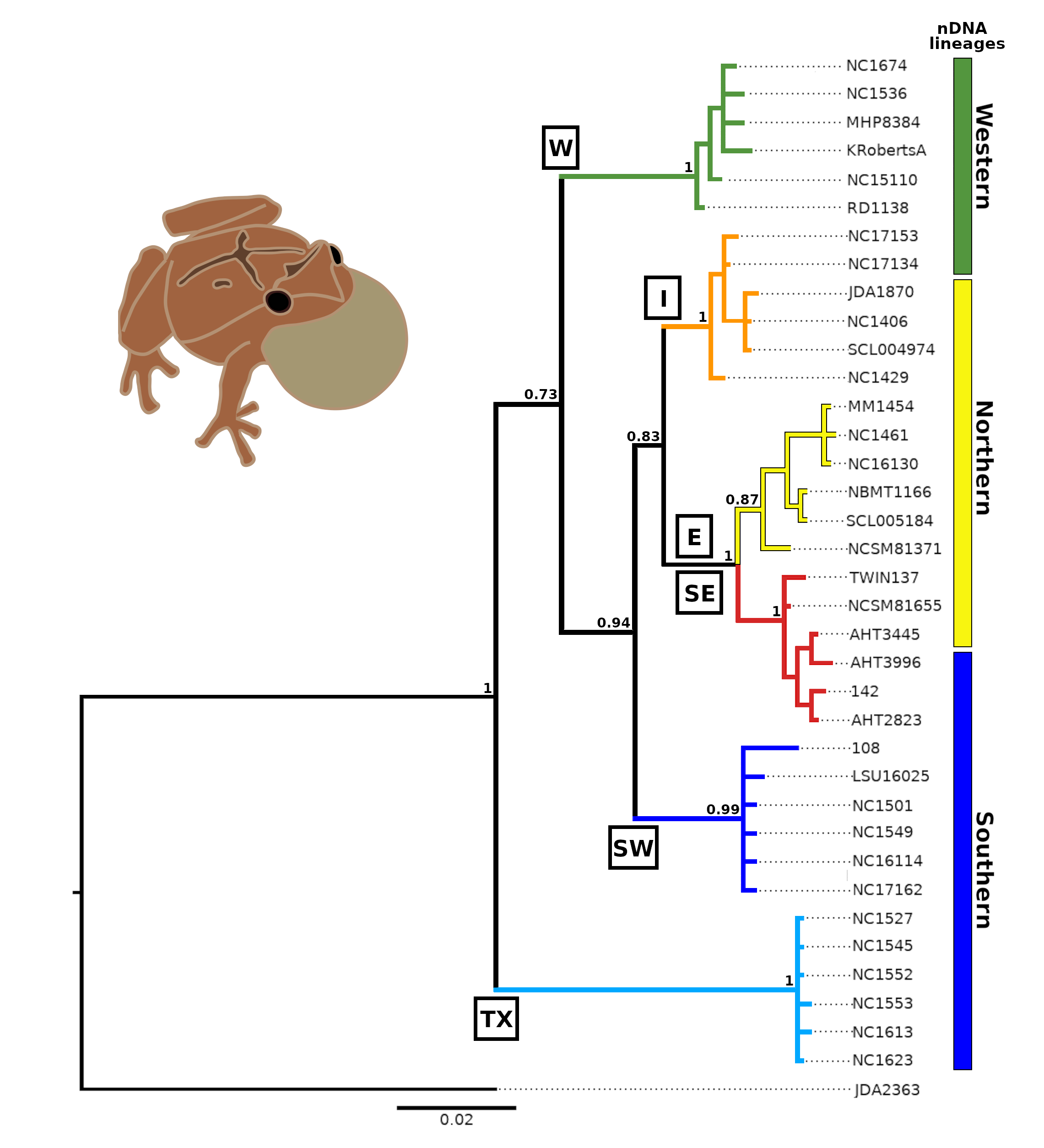
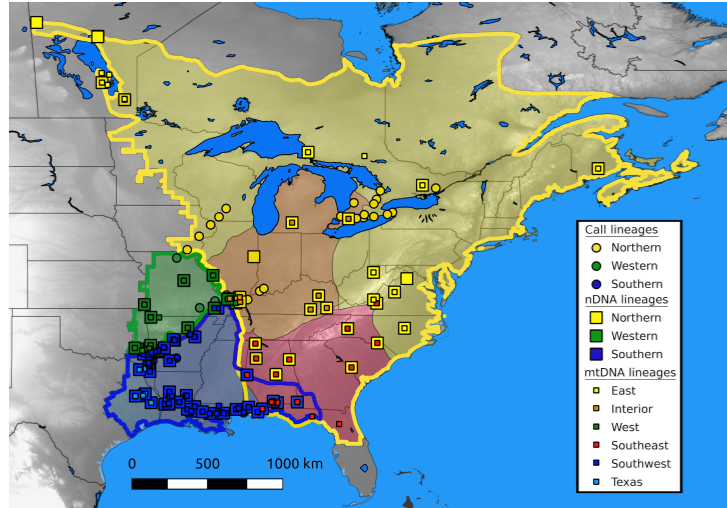
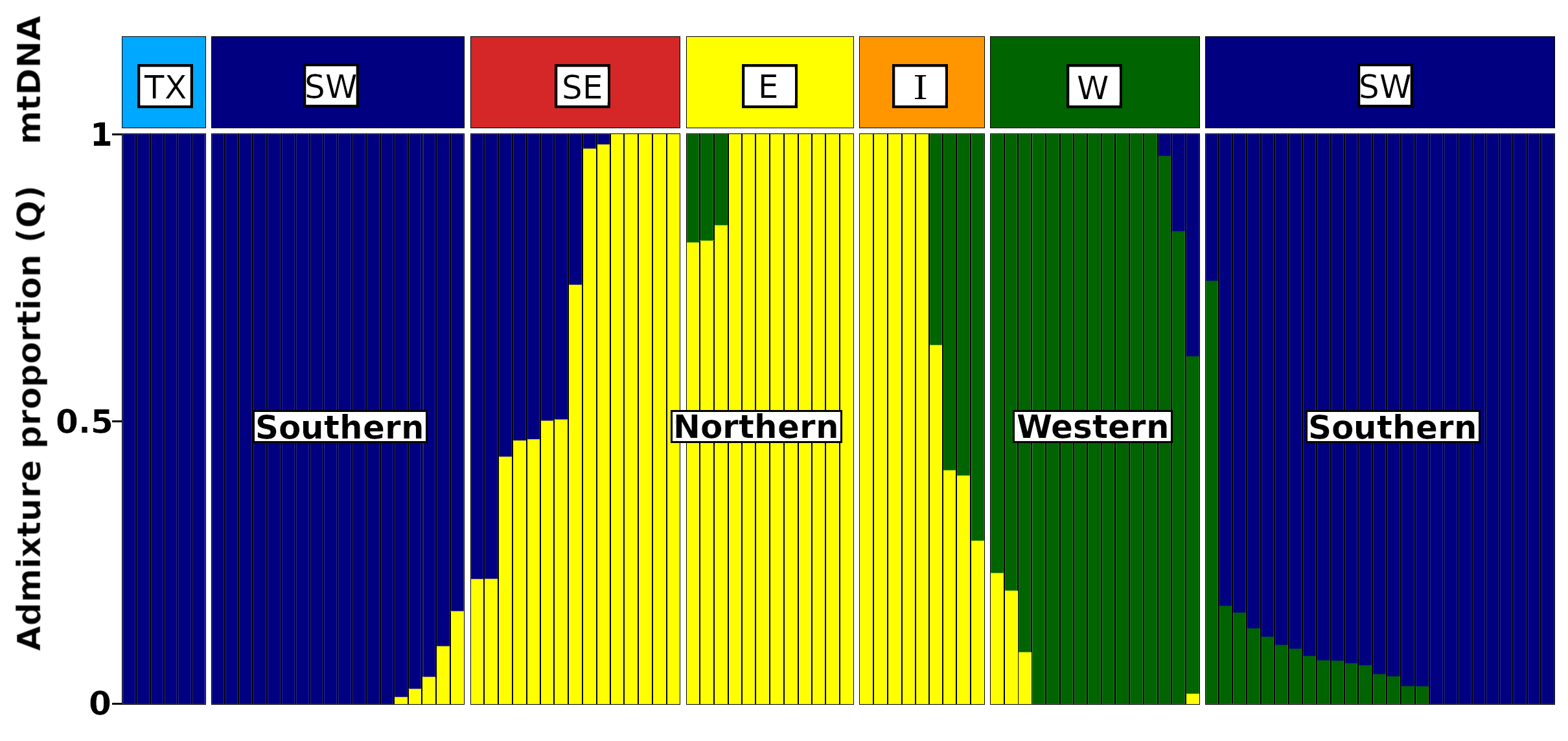
 Among evolutionary biology’s central and most urgent challenges is to explain the distribution of diversity, and nature and timing of the evolutionary processes that shape it. An important facet of my research looks at how historical processes, such as range fragmentation (e.g. glacial refugia), shaped range dynamics, contemporary geographic distributions, and opportunities for hybridization. I combine broad scale fieldwork with phylogeographic and population genetic analyses, tests of mate preference, and in vitro hybridization experiments in a North American treefrog species. The spring peeper (Pseudacris crucifer), one of North America’s most common frog species, has been shown to actually comprise of 6 distinct mtDNA lineages across its geographical range, and these lineages have diverged at various junctures throughout history due to habitat fragmentation (glacial refugial isolation). Upon secondary contact during post-glacial expansion, these intraspecific lineages demonstrate strong prezygotic isolation barriers (divergence in spermatozoa, calls, morphometrics, and strong female preference for natal lineage calls) and hybridization appears maladaptive (cytonuclear discordance within the hybrid zone, unique calls, aberrant morphometrics, and tadpole development). Together, this work suggests the morphologically cryptic lineages of the spring peeper may well be on their way to reproductive isolation through the process of character displacement and avoidance of maladaptive hybridization (reinforcement).
Among evolutionary biology’s central and most urgent challenges is to explain the distribution of diversity, and nature and timing of the evolutionary processes that shape it. An important facet of my research looks at how historical processes, such as range fragmentation (e.g. glacial refugia), shaped range dynamics, contemporary geographic distributions, and opportunities for hybridization. I combine broad scale fieldwork with phylogeographic and population genetic analyses, tests of mate preference, and in vitro hybridization experiments in a North American treefrog species. The spring peeper (Pseudacris crucifer), one of North America’s most common frog species, has been shown to actually comprise of 6 distinct mtDNA lineages across its geographical range, and these lineages have diverged at various junctures throughout history due to habitat fragmentation (glacial refugial isolation). Upon secondary contact during post-glacial expansion, these intraspecific lineages demonstrate strong prezygotic isolation barriers (divergence in spermatozoa, calls, morphometrics, and strong female preference for natal lineage calls) and hybridization appears maladaptive (cytonuclear discordance within the hybrid zone, unique calls, aberrant morphometrics, and tadpole development). Together, this work suggests the morphologically cryptic lineages of the spring peeper may well be on their way to reproductive isolation through the process of character displacement and avoidance of maladaptive hybridization (reinforcement).
- Cairns N, Cicchino AS, Stewart KA, Austin J, Lougheed SC. Cytonuclear discordance, reticulation and cryptic diversity in one of North America’s most common frogs. (in revision)
- Stewart KA, Hudson C, Lougheed SC. 2017. Can alternative male mating tactics facilitate introgression across a hybrid zone by circumventing female choice? Journal of Evolutionary Biology. 30(2): 412-421.
- Stewart, KA, Austin, JD, Zamudio KR, & SC Lougheed. 2016. Contact zone dynamics during the early stages of speciation in a chorus frog (Pseudacris crucifer). Heredity. 116(2): 239-247.
- Stewart KA, Wang R, Montgomerie R. 2016. Extensive variation in sperm morphology in a frog with no sperm competition. BMC Evolutionary Biology. 16(1):29.
- Stewart KA, Lougheed SC. 2013. Testing for intraspecific postzygotic isolation between cryptic lineages of Pseudacris crucifer. Ecology and Evolution. 3(14): 4621-4630.
- Stewart KA. 2009. The Northern Spring Peeper / Rainette Crucifère (Pseudacris crucifer crucifer). Species Account. Opinicon Natural History Notes. Available online Sept. 2009. http://opinicon.wordpress.com/species-accounts/spring-peeper/
 I have similarly studied the initiation of diversification by investigating the causes and consequences of female preference for local male dialects in the song sparrow (Melospiza melodia melodia). Involving the analysis of an individual’s song composition and the genetic relatedness of these individuals to one another (genetic similarity to the population at-large), evolutionary consequences to local dialect preferences points to local genetic adaptation, potentially a stepping-stone to ecological diversification and speciation.
I have similarly studied the initiation of diversification by investigating the causes and consequences of female preference for local male dialects in the song sparrow (Melospiza melodia melodia). Involving the analysis of an individual’s song composition and the genetic relatedness of these individuals to one another (genetic similarity to the population at-large), evolutionary consequences to local dialect preferences points to local genetic adaptation, potentially a stepping-stone to ecological diversification and speciation.
- MacDougall-Shackleton SA, Dindia L, Newman A, Potvin D, Stewart,KA, MacDougall-Shackleton EA. 2009. Songs, stress and survival in sparrows. Biology Letters. 5 (6): 746-748.
- MacDougall-Shackleton EA, Stewart KA, Potvin DA, Tennenhouse E. 2009. The rich get richer: song complexity predicts song element sharing and song output in song sparrows Melospiza melodia. Animal Behaviour 78 (1): 141-146.
- Stewart KA, MacDougall-Shackleton EA. 2008. Local song elements indicate local genotypes and predict physiological condition in song sparrows, Melospiza melodia. Biology Letters 4 (3): 240-242.
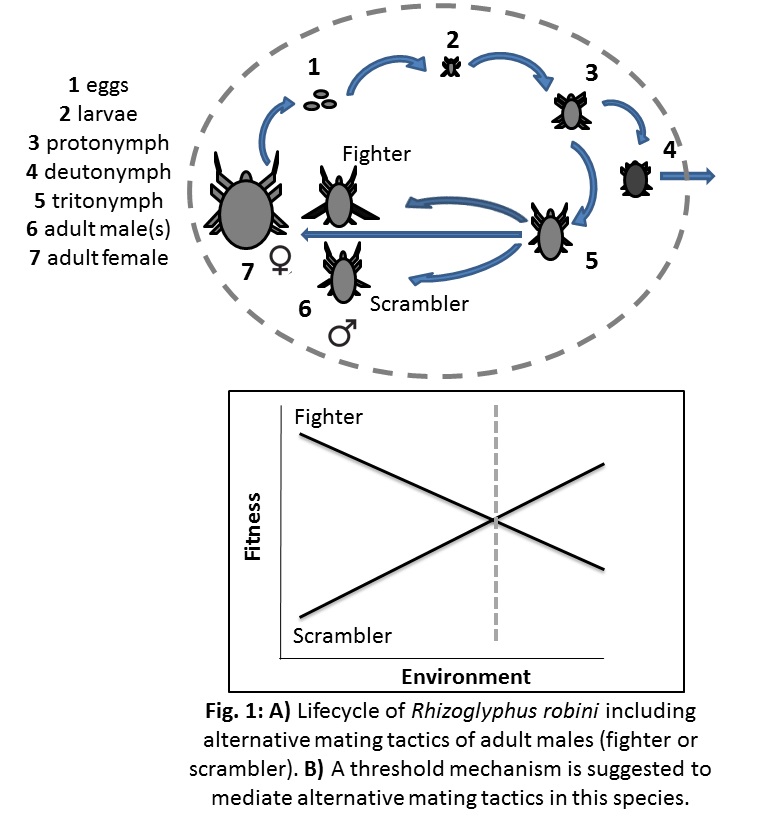 At UvA, I investigate gene-by-environmental interactions, looking at the effects of different ecological and evolutionary processes on population sustainability under environmental change, quantifying the genetic underpinnings of plasticity responses using a cosmopolitan crop pest species as a model. Through the use of experimental evolution and high-throughput sequencing, this research aims to answer questions that bridge topics of dispersal ecology, climate change, and population genetics, such as: 1) Do genetic bottlenecks influence environmental adaptability? 2) Does phenotypic and genetic variance buffer against environmental perturbations and increase population persistence? My model species is the bulb mite (Rhizoglyphus robini), that is well suited for broad-scale experimental evolution projects. Interestingly, this species comprises males with alternative mating morphologies; upon maturation to adulthood, males either morph into a fighter with sharp and thickened third leg pairs used to fight competitors for access to females, and smaller scramblers that lack such armaments. These individuals offer an indispensable opportunity to study alternative mating tactics, and the eco-evolutionary dynamics governing them.
At UvA, I investigate gene-by-environmental interactions, looking at the effects of different ecological and evolutionary processes on population sustainability under environmental change, quantifying the genetic underpinnings of plasticity responses using a cosmopolitan crop pest species as a model. Through the use of experimental evolution and high-throughput sequencing, this research aims to answer questions that bridge topics of dispersal ecology, climate change, and population genetics, such as: 1) Do genetic bottlenecks influence environmental adaptability? 2) Does phenotypic and genetic variance buffer against environmental perturbations and increase population persistence? My model species is the bulb mite (Rhizoglyphus robini), that is well suited for broad-scale experimental evolution projects. Interestingly, this species comprises males with alternative mating morphologies; upon maturation to adulthood, males either morph into a fighter with sharp and thickened third leg pairs used to fight competitors for access to females, and smaller scramblers that lack such armaments. These individuals offer an indispensable opportunity to study alternative mating tactics, and the eco-evolutionary dynamics governing them.
- Smallegange IM, Rhebergen FT, Stewart KA. Cross-level considerations for explaining selection pressures and the maintenance of genetic variation in condition-dependent male morphs. Current Opinion in Insect Science. 36: 66-73. DOI: 10.1016/j.cois.2019.08.005
- Stewart KA, Draaijer R, Kolasa MR, Smallegange IM. 2019. The role of genetic diversity in the evolution and maintenance of environmentally-cued, male alternative reproductive tactics. BMC Evolutionary Biology. 19:58. DOI: 10.1186/s12862-019-1385-4
- Stewart KA, Van den Beuken TPG, Rhebergen FT, Deere JA, Smallegange IM. 2018. Evidence for a third male type in a male-dimorphic model species. Ecology. 99(7):1685-1687
(ii) Novel molecular tools to answer applied questions
Environmental DNA (eDNA) survey methods rely on the presence of DNA fragments that have been shed by organisms into various habitats, and in so doing, have been used to infer species distributions, conservation status (or otherwise health of a community), and applied for ecological surveillance of rare, cryptic or otherwise hard-to-study taxa (e.g. endangered species).
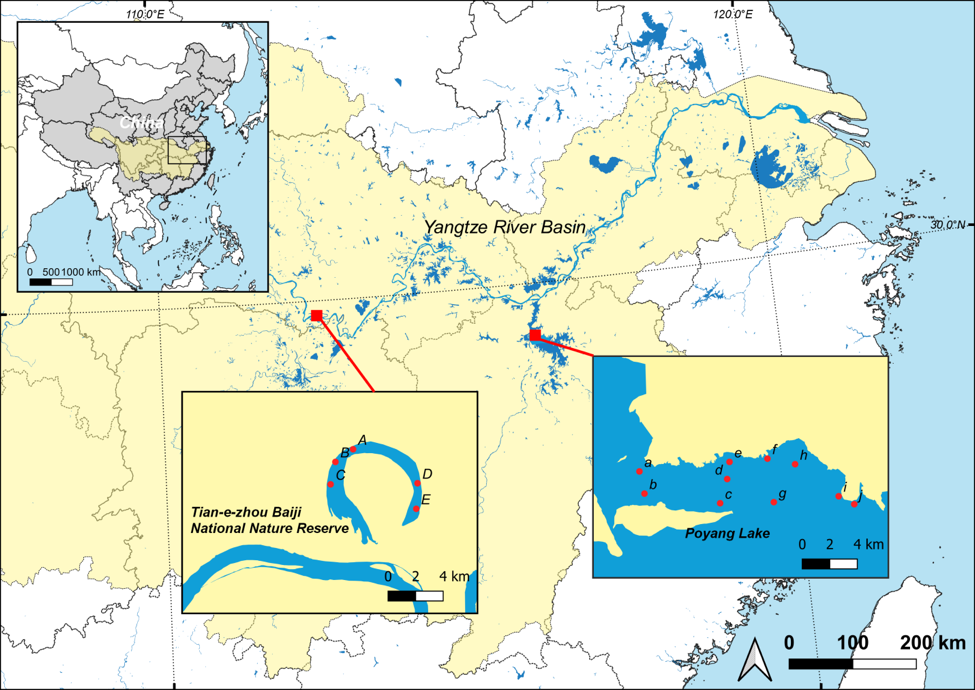
An ongoing research program I have at Tongji University integrates the use of conservation genetics in Chinese policy and management. Utilizing eDNA tools in conserving a critically endangered species, the Yangtze Finless Porpoise (Neophoceana asiaeorientalis), my team and I have been working in conjunction with WWF China to initiate and implement the use of non-invasive sampling to better describe the distribution and ecology of this species throughout the Yangtze River Delta. We use various methods of amplification (PCR, qPCR) and metabarcoding to understand how eDNA can be used to gain insights into spatial distributions, predator-prey dynamics, and trophic interactions.
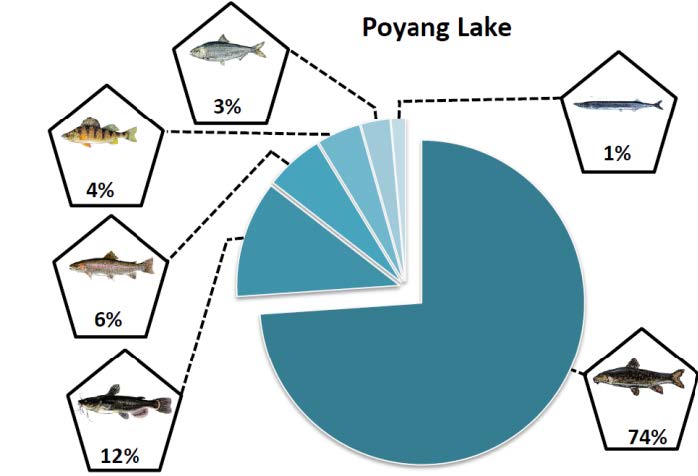
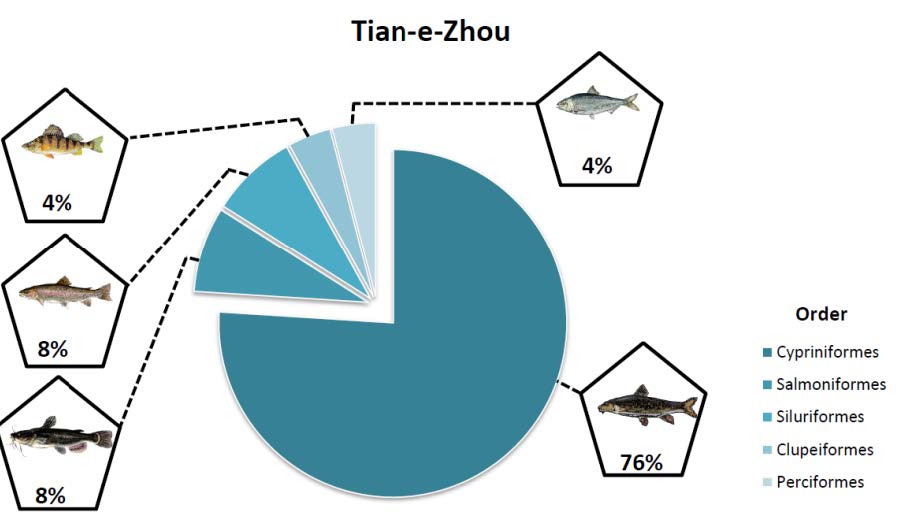
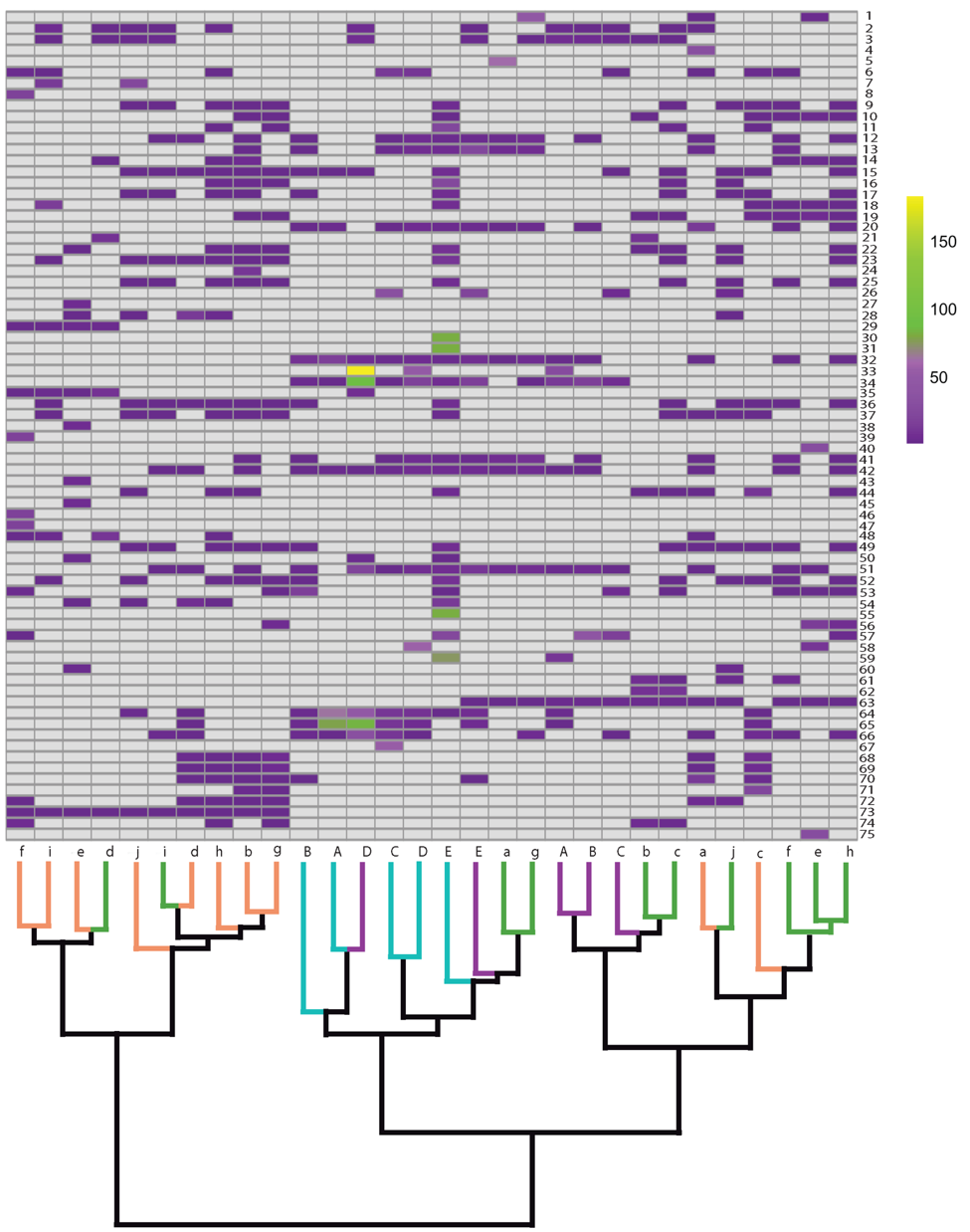
Recently, we have mapped fish community constituents across the Yangtze River Delta, including key prey species of Finless Porpoises, potentially enabling us to draw links between prey distribution and abundance, and endangered population strongholds in nature.
Key research objectives include: 1) Combining data from diverse sources to estimate biodiversity, 2) Detailing how uncertainty in estimates of biodiversity often depends on the types of sampling chosen, and 3) Combining estimates of biodiversity with physical covariates to obtain regional inferences about biodiversity (e.g. predict optimal locations for nature reserves).
- Qu C, Stewart KA, Clemente-Carvalho R, Zheng J, Wang J-X, Gong C, Ma L, Zhao J, Lougheed S. Comparing fish prey diversity for a critically endangered aquatic mammal in a reserve and the wild using eDNA metabarcoding. Scientific Reports. 10: 16715.
- Qu C, Stewart KA. 2019. Evaluating monitoring options for conservation: traditional and environmental DNA tools for a critically endangered mammal. Science of Nature: Naturwissenschaften. 106:9. DOI: 10.1007/s00114-019-1605-1
- Stewart KA, Ma H, Zheng J, Zhao J. 2017. Using environmental DNA to assess population-wide spatiotemporal reserve use. Conservation Biology. 31(5):1173-1182
- Ma H, Stewart KA, Lougheed SC, Zheng J, Wang Y, J Zhao. 2016. Characterization, optimization, and validation of environmental DNA (eDNA) markers to detect endangered aquatic mammals. Conservation Genetic Resources. doi:10.1007/s12686-016-0597-9